Investigator Resources
- Details
- Written by Stephanie Corkran
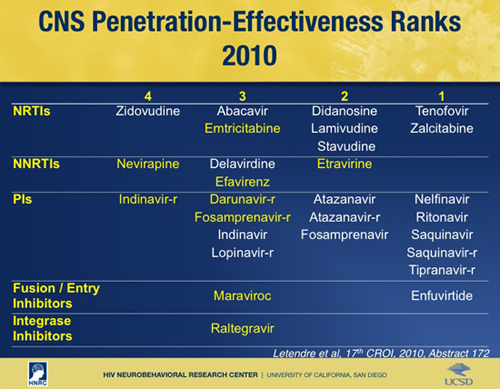
Letendre SL, FitzSimons C, Ellis RJ, Clifford D, Collier AC, Gelman B, Marra C, McArthur J, McCutchan JA, Morgello S, Simpson D, Vaida F, Heaton R, Grant I, and the CHARTER Group. (2010). Correlates of CSF Viral Loads in 1,221 volunteers of the CHARTER cohort. 17th Conference on Retroviruses and Opportunistic Infections.
- Details
Data Management and Information Systems Overview
The Data Management and Information Systems (DMIS) Unit provides HNRP investigators and staff with a state-of-the-art web-based data management and reporting systems, and both the hardware and software for collaborative research and communication.
Organization
The work of the HNRP DMIS unit is directed by, Anthony Gamst, Ph.D. and managed by Clint Cushman. Dr. Gamst assures that the conceptual and practical operations of the DMIS Unit conform to the highest standards and meet the needs of the HNRP investigators. He is responsible for developing the conceptual data management modeling and for assuring the implementation of the data management plan which will use that model to provide flexibility, accountability, and timeliness of data acquisition, storage and access.
As the DMIS Manager for the HNRP, Mr. Clint Cushman, under Dr. Gamst’s direction, oversees the design, development and the use of the HNRP web-based client/server databases. He ensures that, on a day-to-day basis, there is close coordination regarding work for specific HNRP projects. Mr. Cushman directly supervises the programming and technical support staff.
Resources Provided
The HNRP DMIS Unit is focused on developing web-based tools for fast, accurate, and secure data entry; designing, building and maintaining secure, high-availability data systems; and developing tools for the secure tracking, reporting and downloading of data and summaries.
In addition to the data collected in pursuit of specific scientific aims, the HNRP data management system tracks and displays agendas, minutes, reports, publications and other information facilitating scholarly exchange, collaboration, education and training. Collaboration is further facilitated in software through the use of secure content management systems, XMPP/IM services, digital white-boards, and video-teleconferencing solutions. The DMIS Group is focused on providing first-class support to HNRP investigators through testing and development of novel systems.
In order to provide ongoing support for research associated with the long-term longitudinal studies of the HNRP, it has been important to provide utilities to facilitate Reproducible Research. To that end, the DMIS unit has developed a suite of utilities to track data access and associate it with publications and research projects. This data is then stored and can be later accessed or associated with additional data from a date-associated frozen dataset.
DMIS Technical Details
The principal focus of the Data Management Unit’s innovation is creating an infrastructure for distance-independent national and international research collaboration using the Internet. For example using a database-driven approach to dynamic web publication called a ‘portal’ approach, web pages are dynamically constructed via scripts that assemble each page ‘on the fly’ as users click to request various types of information.
The HNRP DMIS Group will continue to build upon the experience and expertise of the existing DMIS staff, leveraging existing HNRP infrastructure to meet future needs. The DMIS Group at the HNRP has nearly 20 years experience in HIV research and related areas. Having supported HNRP associated studies including the HIV Neurobehavior Research Center (HNRC, 5 P30 MH062512-08) and over 80 associated studies including major projects such as the NIDA Program Project (5 P01 DA012065-08) and the CNS HIV Anti-Retroviral Effects Therapy Research (CHARTER) Project (N01 MH22005), the DMIS Group has developed a highly effective infrastructure capable of capturing, validating and distributing data, meeting the diverse needs of investigators in a wide range of clinical and cohort studies.
Systems Security. The local network is protected from the outside world by a firewall, which restricts access based on IP address and service/port. All machines on the local network have up-to-date patches, virus and spy-ware protection. Access to the database is available through secure web-forms only and all communication over the local network runs on the secure sockets layer (SSL). User management tools enforce fine-grained access control based on user (login) ID. Authentication is password-based, with two-factor authentication available, using RSA SecurID tokens or USB keys.
Sharing. Users wishing to access HNRP resources are asked to establish a Resource Account. A User Management tool has been developed for this purpose and registered users have access to standardized reports. To reduce the potential for misunderstanding or inadvertent misuse of particular variables, the on-line, searchable data dictionaries are annotated with 'usage and analysis notes' for key (potentially all) variables.As part of the registration process, users are asked to provide basic contact information and required to assent to a Data Use Agreement. Registered users are also be able to request access to additional data, specimens, and expertise (including subject-matter, data management, laboratory, and statistical support). Access requests take the form of brief proposals to be reviewed for scientific merit, feasibility, and availability of resources. This entire process is managed over the web (through a Resource Request Review tool). We request that data derived from HNRP resources be given back to the program for access by other investigators (perhaps after a short embargo period). To further facilitate data exchange between external investigators, user driven bulk upload utilities have been created to provide an easily accessible method of data sharing.
Data Entry. All projects start with the creation of a searchable on-line data dictionary. Data entry screens, back-end tables, and bulk-upload tools (for laboratory, imaging and other high-throughput data) are generated directly from the dictionary. Bulk upload tools are also available for incorporation of data with essentially arbitrary structure, using an entity-attribute-value (EAV) model for the back-end tables. All data entry screens are accessible via the internet to allow for off-site data entry.
Integrity. The database is fully normalized, reducing the risk of mismatch, synchronization, and other errors. Data warehousing, where necessary, is accomplished by spinning off views. Data is never deleted from the system and all modifications are stored in a transaction log which can be used to roll back to previous dataset states in the event of detected errors.
Quality Control (QC). We have a robust, multi-level data quality control system. Data entry error-rates (on and off-site) have been less than 0.1% (on all items). Double entry is used for on-site quality control. Duplicate records are compared after entry, local QC staff follows-up on and corrects all discrepancies, and the (corrected) record is committed to the database. Logic, range and cross-form checks are then applied to all data and a report on errors and unusual values is sent to staff of the appropriate core or project, returning corrections to DMIS staff. Single-entry data is subject to a 20% random audit, with additional follow-up and training should error-rates rise above 0.1%. Staff experts in each Core are associated with each element of the dataset and are expected to annotate the on-line searchable data dictionary, commenting on issues related to coding, analysis or interpretation of the corresponding values. End-users can use this information to avoid common inferential errors. End-users discovering potential errors in the data are asked to write to the DMIS QC email alias, so that DMIS staff may follow-up with Core experts to resolve the issue. Email describing the resolution will then be sent to the end-user.
Availability. Mission-critical systems are continuously monitored and pages are sent to IT staff on failure. Routers, servers and HVAC equipment are attached to three-hour uninterruptible power supply (UPS). Developers work on hardware identical to that used in production so that components can be swapped on hardware failure, should it occur. We have less than two hours of unplanned down-time per year.
Backup and Disaster Recovery Incremental back-ups are performed nightly. Full back-ups are obtained weekly and stored, in perpetuity, off-site in locked, fire-safe media storage.
IT Support. The information technology support component of the HNRP is responsible for resolving over 6,000 requests per year and maintaining 100+ desktop/laptop machines for staff at multiple locations. The group is also responsible service/deployment of videoconferencing resources and for updates to and security maintenance of web and database servers. During the proposed funding period, the existing data team will be enhanced with a dedicated IT Security specialist who will be responsible for auditing current resources and developing plans for future expansion.
Innovations. Given the diverse requirements of multiple studies, the HNRP DMIS group has developed extremely versatile data structures over its 20 year history. By using an event-based data management system, the DMIS Unit provides a structure, which allows for events to be defined on-the-fly and accommodates both cross-sectional and longitudinal data. This addresses long-standing issues of database flexibility for long-term cohort studies. The event based nature of this structure allows us to adapt to changing circumstance, interest, and scientific focus without having to completely re-write the data system. This structure combined with our robust protocol-based data dictionary allows for personalized case-report forms and easy integration of data across multiple protocols.
A recent development has been a conversion from trigger-based calculations combined with stored summary scores to dynamic calculations on the fly to allow for scoring algorithm version control and easy comparisons to earlier results. This change allows us to look at newly collected data alongside older results easily using common algorithms as well as reducing the processor overhead at time of entry.
Flexibility. A specific example of flexibility in data structure is our emphasis on Entity Attribute Value (EAV) structures for external data. The EAV structure allows for flexibility in storage (for example, of variable length lists), and allowed us to develop web-based upload tools to allow off-site investigators and future external investigators to define, upload and report on data (such as imaging or neuropathology results) they provide to support HNRP projects.
Reporting. DMIS has created several web-based utilities to support multi-site collaborations. These utilities range from dynamically generated web based cohort growth charts/reports to interactive enrollment, scheduling and cohort/specimen tracking utilities. To accommodate the needs of HNRP investigators, all data tables have been made available for download via a web-based data archiving utility. Additionally, investigators are provided resources to store queries to generate project-specific datasets on the fly.
Collaboration. To facilitate communications among investigators who are physically dispersed (e.g. imaging resources are on La Jolla UCSD campus while the HNRC resides on Hillcrest campus), we coordinate as much as possible via the web to reduce the requirement for numerous face-to-face meetings, while emphasizing close communication. To that end, we have deployed a wiki-based content management system for all study web resources to provide HNRP investigators and staff direct control of content, Instant Messaging (IM) services for study staff and digital white-boards/video-teleconferencing for cross-site meetings. As an example it is possible for Participant Unit staff to directly interface with the participant community by providing health links and recruiting information for the various projects as new information comes to them. Additionally, project personnel are able to interact directly with support staff within the administrative core via IM and web-based meeting resources to help maintain a cohesive center.
Contact
For assistance, please contact Clint Cushman (DMIS Manager) at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
Laboratory, Pharmacology & Biomarkers Resource (LPBR) Overview
The LPBR oversees the laboratory components of HNRP-associated projects. Depending on the needs of each individual project, the LPBR collects, processes, stores and makes available for future research, samples of plasma, serum, CSF and CSF cell pellets, urine, and genetic material (e.g. PBMCs). The LPBR coordinates routine clinical laboratory testing of these samples and also provides the HNRP with the capacity to measure ARV medication concentrations, immune mediators, and other biomarkers in CSF and blood. This group also facilitates genetics and genomics studies. Although this work is primarily performed on samples from human research participants, the group also performs some work using animal samples.
Organization
The LPBR is housed within the Neuromedical Resource Group (NMRG). The same leadership team directs the work of the LPBR and NMRG: Dr. Ronald Ellis (NMRG Director), a neurologist with subspecialty expertise in neurobehavior and HIV/AIDS; Dr. Scott Letendre (Co-Director), an infectious disease specialist with expertise in neuroAIDS and Hepatitis C infection; Terry Alexander (NMRG Manager), a clinical research nurse with over ten years of experience in applying neuromedical research methods in local and international settings. Within this NMRG structure, Debra Rosario, MPH, directly manages LPBR activities and staff, including laboratory assistants and research associates trained in phlebotomy and handling biological specimens.
Resources provided
LPBR resources include:
- Personnel trained in the collection and processing of participant-derived samples.
- Laboratory data already collected from HNRP-associated projects (with consents for data sharing) and the ability to obtain new laboratory data through retrospective testing of existing specimens or collection and testing of new specimens.
- Storage and management of more than 500,000 specimens collected from subjects with comprehensive neuromedical and neuropsychological data who have participated in HNRP-associated studies.
- Substantial experience in measuring and interpreting biomarker and pharmacology concentrations in relationship to HIV, methamphetamine abuse and other related conditions.
Scientific Consultation and Trainees
Although scientific consultation and training activities may be laboratory specific, these activities are addressed at the larger NMRG level.
Cross-project Issues
In addition to using NMRG methods for addressing cross-project issues, the LPBR subgroup meets weekly to address all laboratory issues including standardization of assays and methodologies across projects. A meeting focused on pharmacology issues takes place monthly.
Contact the LPBR
For additional information or to access LPBR resources, please contact LPBR manager, Debra Rosario (This email address is being protected from spambots. You need JavaScript enabled to view it.).
LPBR Technical Details
General Procedures
Processing of Participant-Derived Blood and CSF Samples. Specific processes depend upon the details of each research protocol. But in general blood and CSF are collected, processed and separated, and aliquotted for storage or transfer to multiple clinical and research laboratories. Blood, collected by phlebotomist, is sent to UCSD clinical laboratories or Lab Corp (San Diego, CA) for hematology, chemistry, T-lymphocyte subset enumeration by flow cytometry, and HIV RNA quantitation by Roche PCR ultrasensitive assay (lower limit of detection 50 copies per mL). CSF,collected by the physicians or nurses, is sent to UCSD labs for cell count, glucose and total protein measurements, and HIV RNA quantitation. Both blood and CSF can be assayed in-house for ultrasensitive HIV RNA quantitation by Abbott PCR ultrasensitive assay (lower limit of detection 40 copies per mL), rapid screenings of HIV and HCV (Medmira, Nova Scotia, Canada), syphilis (RPR), urine toxicology, and specialty assays such as measurements of drug and biomarker concentrations. Urine specimens are screened for neuroactive drugs at the time of cognitive testing by a 10-panel drug screen card (Rapid Response, Biotechnostix Inc., Markham, Canada) that immediately detects exposures to recreational or prescription drugs.
|
Table 1. Typical sample volumes stored by at each HNRP-associated study visit. Volumes may vary based on the requirements for each study supported by the NMRG. |
|||
|
Sample Type |
Aliquot size x Number |
Approx. Total |
Temp. Stored |
|
Serum |
1.0 x 5 |
5 mL |
-20°C |
|
EDTA Plasma |
1.0 x 10 |
10 mL |
-70°C |
|
PBMCs / DMSO |
1.8 x 2 |
3.6 mL |
-150°C |
|
CSF Supernatant |
0.5 x 6 |
3 mL |
-70°C |
|
CSF Supernatant |
1.0 x 5 |
5 mL |
-70°C |
|
CSF Cells |
0.5 x 3 |
1.5 mL |
-150°C |
Labeling and Tracking. The LPBR carefully tracks all processed specimens. Barcode labels identify precise freezer locations (freezer number, rack number, box number). These codes are tracked by a web-based data entry program, which was designed and developed by the HNRP’s Data Management and Information Systems Resource. Entry of sample data by barcode also facilitates automatic and immediate update of the fluids database; accounting of sample use when a sample is processed or retrieved; and prioritization and appropriate utilization of samples. Disposition of stored fluid samples is readily defined by this tracking system. When aliquots are required for research assays or shipment to collaborators, samples are logged out and the recipient recorded. If a previously removed sample is returned after partial use, it is assigned a new location and its history noted in the database. Thus, this system enables tracking of the history of individual samples.
Specimen Storage. The HNRP Specimen Storage Facility is comprised of -20°C, -70°C, and -150°C freezers that house all HNRP-associated biological specimens. These freezers are monitored 24 hours a day via a computerized validated environmental monitoring system that is capable of automatically phoning key personnel in the event of a power failure or freezer mechanical failure. All freezers have a posting on them listing personnel to call in the event of an emergency. The Standard Operating Procedures outline steps to take in the event of a power failure and/or freezer mechanical failure. All staff and Principal Investigators have been trained to respond to the freezer alarm system and have been given instruction of how to proceed if there is a situation requiring immediate action. Coverage is always designated to account for vacations and holiday schedules.
Participant identification, specimen procurement, labeling and confidentiality. Laboratory or clinical personnel verify the subject’s name, study identifier, visit, and date prior to procuring fluids. Local site personnel clearly label the remaining specimens with this unique identifying information prior to freezing.
LPBR personnel performance. Copies of LPBR personnel training records are maintained. Laboratory personnel record processing times for the fluids obtained from each subject on clinical report forms and monitor these forms periodically to ensure that processing times are within 2 hours from procurement to either shipment or freezing. Efforts are made to avoid unnecessary thawing of specimens and prolonged periods at room temperature since they may adversely affect the integrity of the constituents of biologic samples.
Laboratory instrumentation, reagents, and analytic test procedures. Each clinical lab utilized by the LPBR, whether local or central, is accredited by the College of American Pathologists and or Clinical Laboratory Improvement Amendments. The designated, commercial lab is responsible for maintaining its instrumentation, reagents, and test procedures. UCSD’s Environmental Health and Safety Office monitors intramural research reagents and test procedures. Each lab verifies that they are maintaining calibration and accuracy of their instrumentation.
Turnaround times. Clinical lab results are delivered by the central, commercial lab in electronic format to the LPBR, facilitating easy monitoring of turnaround time.
Accuracy of final results. When aberrant lab results are found, their occurrence is tracked. LPBR personnel also track when such results require repeat processing. Errors are reviewed annually to identify problems with specific assays or procedures and performance of the central, commercial lab.
Complete documentation of all procedures involved in obtaining final analytic results. Documentation is maintained and monitored in a systematic manner by the central lab in the process of QA, it is important to document the effectiveness of quality control measures. The written record of quality control activities for each procedure or function should also include details of deviation from the usual results, problems, or failures in functioning or in the analytic procedure, as well as any corrective action taken in response to these problems.
Additional measures to support quality of processing and uniformity results will include:
- When thawing is necessary for processing, residual volumes that are returned to storage are marked as previously thawed and a notation will be recorded in the database.
- The accuracy of the specimen database is continually tested as groups of samples are withdrawn for specific assays. Inconsistencies (i.e., failure to find a sample in its stated location) are reported and the database promptly corrected.
- Developmental and Pilot Studies. The NMRG maintains a record of pilot studies supported, funded grants utilizing, and publications emerging from provided samples
Laboratory Operations Manual
The LPBR has developed a Laboratory Operations Manual as a resource for Laboratory Assistants and Clinical Research Assistants that includes general policies and guidelines pertaining to personnel, standard operating procedures and QA. Specifically, the Operations Manual highlights key elements of the Biomarkers Laboratory with regard to Assay Protocols, Data Collection and Analysis, Business and Administration and an extensive QA Program that includes validation, certification and evaluation of procedures, equipment and personnel.
Biomarker Quality Assurance (QA)
The Biomarker QA program incorporates several elements, including verification of specimen labeling; tracking of laboratory personnel performance; certification of laboratory instrumentation, reagents, and analytic test procedures; assessing precision and accuracy of final results; entry of final results into the central database; and complete documentation of all procedures involved in obtaining final analytic results. To assess precision of our methods, we assay specimens in duplicate, repeating measurements that have coefficients of variation greater than 20% (~5% of assays), and repeating 10% of all biomarker assays to assess operator and batch consistency.
- Details
Developmental grants are funded through the Developmental Core of the HNRC.
The primary goal of the Developmental Grants Program (DGP) will be the initiation of innovative studies by junior faculty and trainees at UCSD or affiliated institutions with the following objectives:
- Recruitment to neuroAIDS research of new investigators or established investigators without prior experience in the field;
- Generation and pilot testing of new research initiatives;
- Fostering collaboration among investigators from throughout Southern California.
The program provides to qualified investigators and trainees any appropriate combination of the following forms of support:
- Small, 1-2 year grants to support pilot studies;
- Access to HNRC core resources such as data, specimens, participants, equipment, administrative support, or expert consultation and technical assistance.
The application and evaluation processes for grants is semiannual, rapid, and relatively simple. For faculty, assistance will be directed to projects that can generate pilot data that support peer-reviewed proposals to outside agencies. For postdoctoral fellows, graduate students, or other trainees, the objective may be completion of projects that fulfill requirements for advanced degrees and/or result in peer-reviewed publications.
Southern California and specifically San Diego hosts a remarkable concentration of talented investigators in the biomedical and neurobehavioral sciences relevant to neuroAIDS. Fostering collaboration among these investigators by providing funding for pilot studies and support for trainees is an important secondary objective of the core.
For more information, click here.
- Details
Neuromedical Resource Group Overview
The Neuromedical Resource Group (NMRG) serves as a resource to HNRP’s associated studies and collaborating investigators. It provides comprehensive clinical and laboratory evaluations as well as scientific consultation, training, and community education in NeuroAIDS. The HNRP’s collaborating investigators find data coordinated through the NMRG (e.g. antiretroviral (ARV) treatment information, HIV disease staging information) to be essential for their data analyses and publications. By assisting investigators in widely diverging disciplines at all levels of project development, the NMRG encourages the application of multiple scientific perspectives and stimulates interdisciplinary collaboration, coordination and dissemination of results.
Organization
The NMRG is comprised of two interacting subunits. The Clinical Research Support Unit (CRSU) utilizes a new model in which evaluations are performed by a team of staff research associates, nurses, nurse practitioners and physicians. Depending upon the needs of the specific project, the CRSU implements methods for structured neuromedical history-taking, neurological and medical assessment and diagnostic algorithms that facilitate establishing diagnoses of central and peripheral complications of HIV, with special emphasis on neurocognitive diagnoses. The Laboratory, Pharmacology, and Biomarker Unit (LPBU) collects, processes, stores, and makes available for future research, samples of plasma, PBMCs, CSF, and for some studies other fluids and tissues such as urine, CSF cell pellets, and buccal swabs. The LPBU also provides the HNRP with the capacity to measure ARV medication concentrations, immune mediators, and other biomarkers in CSF and plasma, and can facilitate genetics and genomic studies. Together these units perform evaluations on HIV+ subjects and HIV- controls.
Leadership functions of the NMRG are vested in a multidisciplinary team of investigators and staff: Dr. Ronald Ellis (NMRG Director), a neurologist with subspecialty expertise in neurobehavior and HIV/AIDS; and Dr. Scott Letendre (Co-Director), an infectious disease specialist with expertise in neuroAIDS and Hepatitis C infection. Management of the NMRG is provided by Terry Alexander, a clinical research nurse with over ten years of experience in applying neuromedical research methods in local and international settings.
Resources provided
The NMRG coordinates the collection of essential clinical data on human study participants, including neuromedical histories, clinical examinations, and laboratory data. A strength of the NMRG is the standardization and depth of the clinical characterization it provides. The Core Assessment is administered to most studies utilizing neuromedical resources and is designed to keep pace with new knowledge and the evolving objectives of current and future HNRP-associated studies. Core assessments offered include neuromedical history, lumbar puncture, urine toxicology, CD4 count and viral load. In addition to the Core Assessment, neuromedical data is collected as needed to meet the aims of specific studies, i.e.,stroke risk calculator, research biomakers in blood and cerebral spinal fluid
Scientific Consultation
A multidisciplinary group comprised of a neurologist, a neuropsychologist, an internist, a nurse-clinician and a psychiatrist meet regularly to assign neurocognitive diagnostic classifications to research participants. Neuropsychological test data and findings from the neuromedical evaluation (Diagnostic Worksheets) are combined to assign the following diagnoses utilizing research diagnostic guidelines: (1) neurocognitively normal, or (2) asymptomatic neuropsychological impairment (NPI), or (3) mild neurocognitive disorder (MND), or (4) HIV associated dementia.
Trainees
The NMRG offers two types of training: 1) preparation of students in a career focused on neuroAIDS 2) instruction of standardized assessments for clinical and laboratory staff assisting with local, multi-center and international studies.
Training is provided to, pre- and post-doctoral trainees, and fellows who will have the opportunity to gain experience in neuromedical, clinical assessments and pharmacology laboratory methods. Drs. Scott Letendre and Ronald Ellis serve as mentors to trainees.
Cross-project issues
The NMRG holds a weekly meeting to discuss progress of ongoing and upcoming projects. This meeting is attended by Drs. Ronald Ellis and Scott Letendre, NMRG manager Terry Alexander, physicians, nurse practitioners, research associates, and laboratory personnel. Attendees are also involved in an interactive inservice presentation to discuss HIV neurologic disorders, participant case presentations, and preliminary study results. Additionally, clinicians and SRAs have regularly scheduled meetings to address specific issues relevant to the success of daily activities within the NMRG.
Neuromedical Resource Group: Technical Details
General approaches/methods
Although it depends upon the specific needs of the study, the core evaluation includes a neurological and general physical examination performed by a clinician (physician, nurse practitioner, nurse) to detect clinical abnormalities at baseline and subsequent visits (if applicable). This examination includes an evaluation of mental status, cranial nerves, motor, sensory, cerebellar function, reflexes and gait. Other evaluations such as gynecological health history questionnaire, anthropomorphic measurements, Unified Parkinson’s Disease Rating Scale, Unified Huntington’s Disease Rating Scale, neuropathy score are performed according to specific study protocols. Training is available through a variety of modalities such as manuals, videotapes, web-based tutorials, seminars, and workshops.
Methods Training
The NMRG provides structured training for physical and neurological exams, lumbar punctures, CRF completion, participant interviews, and study protocol. Training is also provided for processing lab specimens and assays.
Quality Assurance
A weekly Continuing Education In-service program is vital to quality assurance to provide the NMRG with up to date knowledge in areas relevant to research supported by the HNRP. Data is monitored regularly to ensure that data is consistently processed and made available in the database as studies are accrued. CRFs are reviewed for accuracy, logic, and completion prior to submission to data management. Ongoing QA is also focused on certification of laboratory instrumentation, reagents, and analytic test procedures; as well as assessing precision and accuracy of final results.








